Larch (Larix spp.) is widely distributed in temperate mountainous areas, cold temperate plains and alpine climate areas in the northern Hemisphere. It is the absolute dominant tree species in boreal forest (taiga). The total area of larch in China is 10.8351 million hectares, with a plantation area of 3.1629 million hectares, ranking fourth among dominant tree species in artificial coniferous plantations nationwide. Larch is an important timber and ecological species in both the northern and southern subalpine regions of China. Since the 1970s, the research team led by Prof. Zhang Shougong, Academician of the Chinese Academy of Engineering, has been dedicated to the genetic improvement and selective breeding of larch. After achieving the comprehensive renewal of larch breeding generations, the team further implemented the “Whole Genome Sequencing of Larch” project to expand the breadth and depth of their research.
Based on the data of third-generation sequencing (1.30 Tb) and second-generation sequencing (0.52 Tb), the team successfully assembled the larch genome, which is 10.97 Gb in size. With a Contig N50 of 1.09 Mb, they annotated 45,828 protein-coding genes. It was revealed that of the genome, 66.8% consists of repeat sequences, of which LTR-RT makes up 69.86%. An analysis of genomic evolution indicated that the species divergence between larch and Douglas fir occurred approximately 66 million years ago, after which 1,139 gene families expanded, while 581 gene families contracted.
From a 31-year-old full-sibling family of larch, the team selected two groups with significantly different lignin content. Through population transcriptomics, they found that the variation in lignin content in larch is mainly determined by the process of lignin monomer polymerization. Moreover, the expression levels of six genes (LkCOMT7, LkCOMT8, LkLAC23, LkLAC102, LkPRX148, and LkPRX166) showed a significant positive correlation with lignin content.
This study accomplished a high-quality whole-genome map of larch, identifying the critical roles of two genes, LkLACs and LkPRXs, in the formation of wood properties in coniferous trees. It effectively enhanced the continuity of larch research from fundamental to applied studies, laying a foundation for decoding the unique biological features of larch and genome-assisted breeding, providing insights for answering other biological questions of coniferous tree species. The findings have been published in the Journal of Integrative Plant Biology.
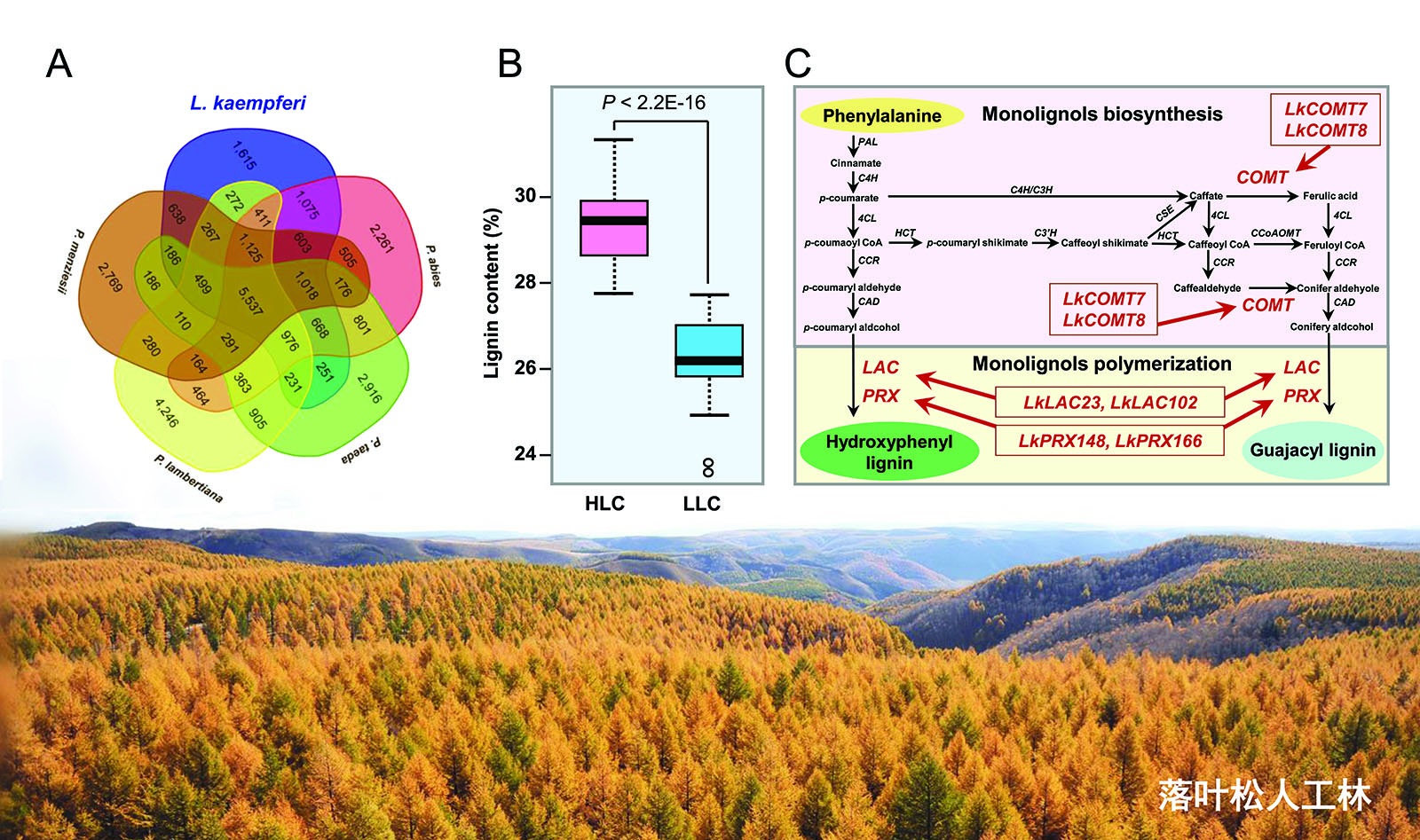
Mechanism of Larch Lignin Monomer Polymerization Regulation